Support Open Science
Accelerate Innovation
Join us in advancing open computational science and making advanced research tools accessible worldwide.
Pitch Deck
For best results, use Google Chrome. Other browsers may not capture slides correctly.
Scientific Computing for Molecular Science Solutions
Breaking Down Complexity for Innovation
From Complexity to Computability
Our platform transforms molecular complexity into manageable computational workflows, enabling scientific breakthroughs through accessible, scalable, and reproducible methods.
$40B+
Scientific Computing (HPC) Market Size (2024)
10-20%
of global supercomputing resources dedicated to molecular simulations
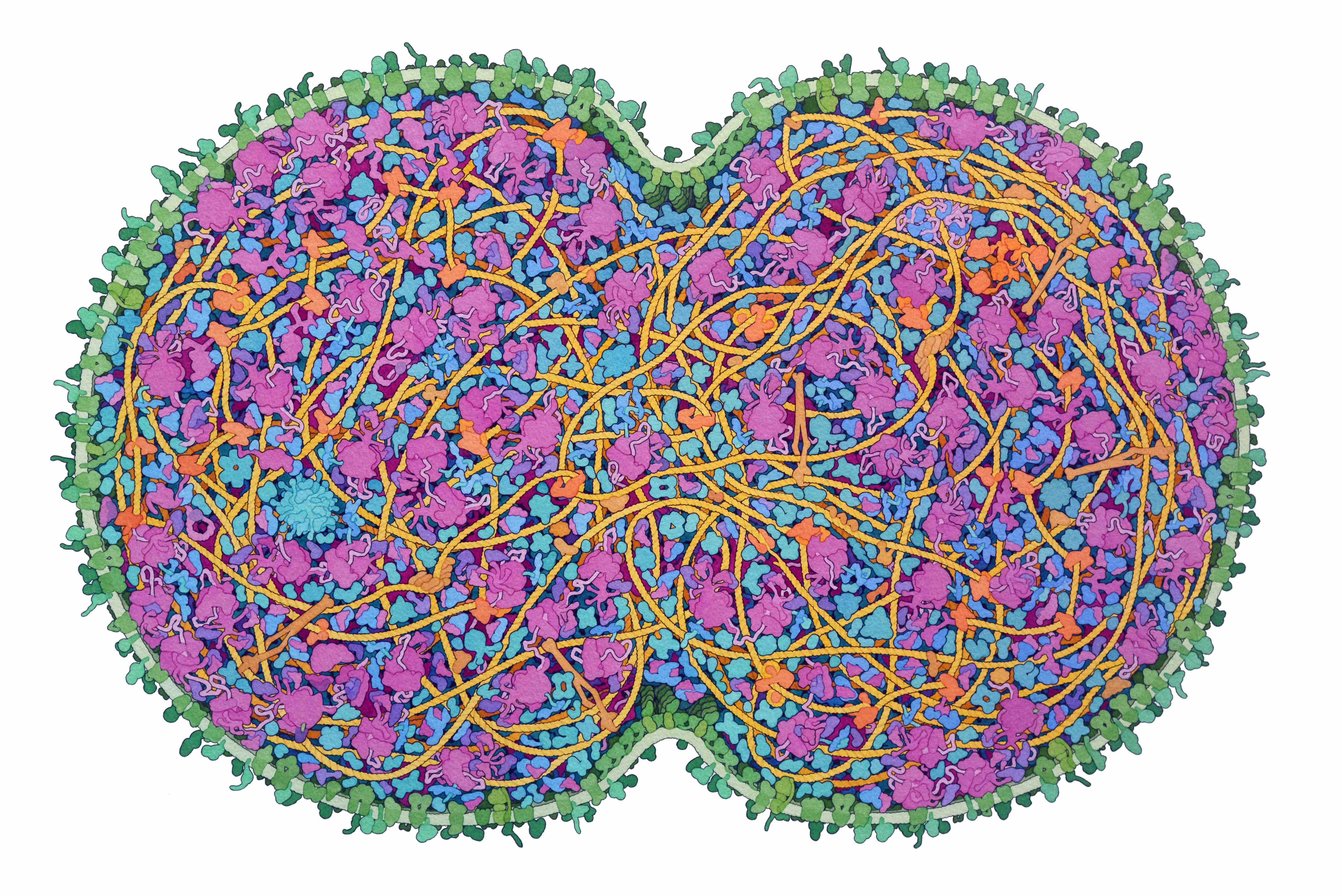
Artistic conception of a cross-section through a dividing minimal cell
RCSB Protein Data Bank. doi: 10.2210/rcsb_pdb/goodsell-gallery-042
Scientific Computing at a Crossroads
The Integration Gap
Researchers spend 60-80% of their time on workflow setup rather than science. The current scientific computing landscape suffers from fragmentation and isolation, creating significant barriers to research efficiency, reproducibility, and knowledge transfer. This leads to the open workflows gap identified in our solution.
- •Licensing costs with declining ROI
- •Proprietary architectures resistant to customization
- •Insufficient methodological transparency
- •Limited data interoperability and integration
- •Complex deployment requirements
- •Overengineered systems with excessive complexity
- •Prohibitive learning curves limiting adoption
- •Lack of standardization across institutions
- •Development driven by immediate project needs
- •Inadequate documentation and maintenance
- •Narrow scope preventing broader application
- •Knowledge silos impeding collaboration
The Giant Missing Piece: Open Workflows
Bridging the Scientific Reproducibility Gap
Despite significant advances in open science infrastructure, a critical component remains underdeveloped: reproducible, reusable computational workflows. This missing element creates barriers to scientific validation, collaboration, and knowledge transfer. BoCoFlow addresses this gap by providing the first comprehensive platform for open workflows.
Current Component
- Established data repositories (PDB, GenBank, materials databases)
- Mature open-source scientific softwares
- Widespread open access publication platforms
- Standardized data formats and exchange protocols
The Critical Gap
- Reproducible computational workflows with full provenance
- Cross-platform execution environments for methodological consistency
- Standardized simulation protocols for result validation
- Integrated knowledge transfer mechanisms for computational methods
"Unfortunately, these rich and costly data are not systematically maintained, and when further analyses are required, simulations have to be rerun—an unacceptable situation from scientific, environmental and sustainability standpoints."
- The need to implement FAIR principles in biomolecular simulations, 2024BoCoFlow: Solving the Open Workflows Gap
Visual Programming Platform for Reproducible Scientific Computing
Addressing the critical gap in open workflows: BoCoFlow transforms complex computational tasks into drag-and-drop nodes, enabling researchers to create reproducible, shareable workflows that reduce setup time by 80% while ensuring full scientific reproducibility.
Cross-Platform Architecture
Three-layered architecture ensuring data security and high performance
Python-Native Node Design
Transform existing code into workflow nodes with minimal modification
Comprehensive Workflow Capabilities

Early Adoption Success Stories
Real-world applications of BoCoFlow in computational chemistry
Streamlining Homology Modeling and MD Simulation Workflows
Research for real-time nucleosome modeling
Manual execution of multiple scripts across different environments. Error-prone process requiring deep expertise in both modeling tools and workflow management.
Automated, visual PdbMDAuto workflow connecting Modeller and GROMACS, allowing non-experts to perform complex modeling with built-in validation checks and reproducibility.
• 80% reduction in workflow setup time
• Elimination of common file format errors
• Ability to easily rerun and modify simulations
"BoCoFlow transformed our homology modeling process from an expert-only task to something our entire team can reliably perform."
The BoundaryComputing Ecosystem
An Integrated Platform for Computational Science Innovation
BoCoFlow
Visual workflow platform for reproducible computational science
Available NowBoCoHub
Repository of verified methods, workflows, and computational modules
Q3 2025BoCoDB
Curated database for scientific discovery and ML applications
Q1 2026BoCoEval
Benchmarking and evaluating computational methods & workflows
Q3 2026BoCoFlowAgent
AI-agent for scientific discovery using workflows
Q1 2027MMarket Expansion Strategy
RKey Revenue Streams
GGrowth Milestones
Meet the Founder
Bringing scientific computing expertise to workflow challenges
"Grounded in firsthand computational science experience, our mission is to transform how researchers build, share, and reproduce scientific workflows."
Supporting Open and Reproducible Scientific Computing
Collaboration with BoundaryComputing
BoundaryComputing builds tools for transparent computational research
Institutional Support
Connect with us through grants and institutional programs
Business Investment
Help us grow through meaningful business relationships
Research Collaboration
Join us in developing solutions for scientific computing
Your Support Makes a Difference
Your contribution helps us develop and maintain open-source scientific tools, provide research grants, and make advanced computational methods accessible to researchers worldwide.
Why Support Us?
- •Accelerate the discovery of new therapeutics and materials
- •Enable broader access to advanced computational methods
- •Support an open ecosystem that benefits researchers worldwide